Rows: 100
Columns: 10
$ Patient_ID <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,…
$ Age <dbl> 71.3, 42.2, 31.9, 38.2, 89.8, 56.5, 87.0, 18.2, 48.1, …
$ Gender <dbl> 0, 0, 1, 0, 1, 0, 0, 1, 1, 1, 0, 1, 0, 0, 1, 1, 0, 1, …
$ Enrollment_Date <date> 2022-11-12, 2022-12-16, 2022-11-20, 2022-10-05, 2022-…
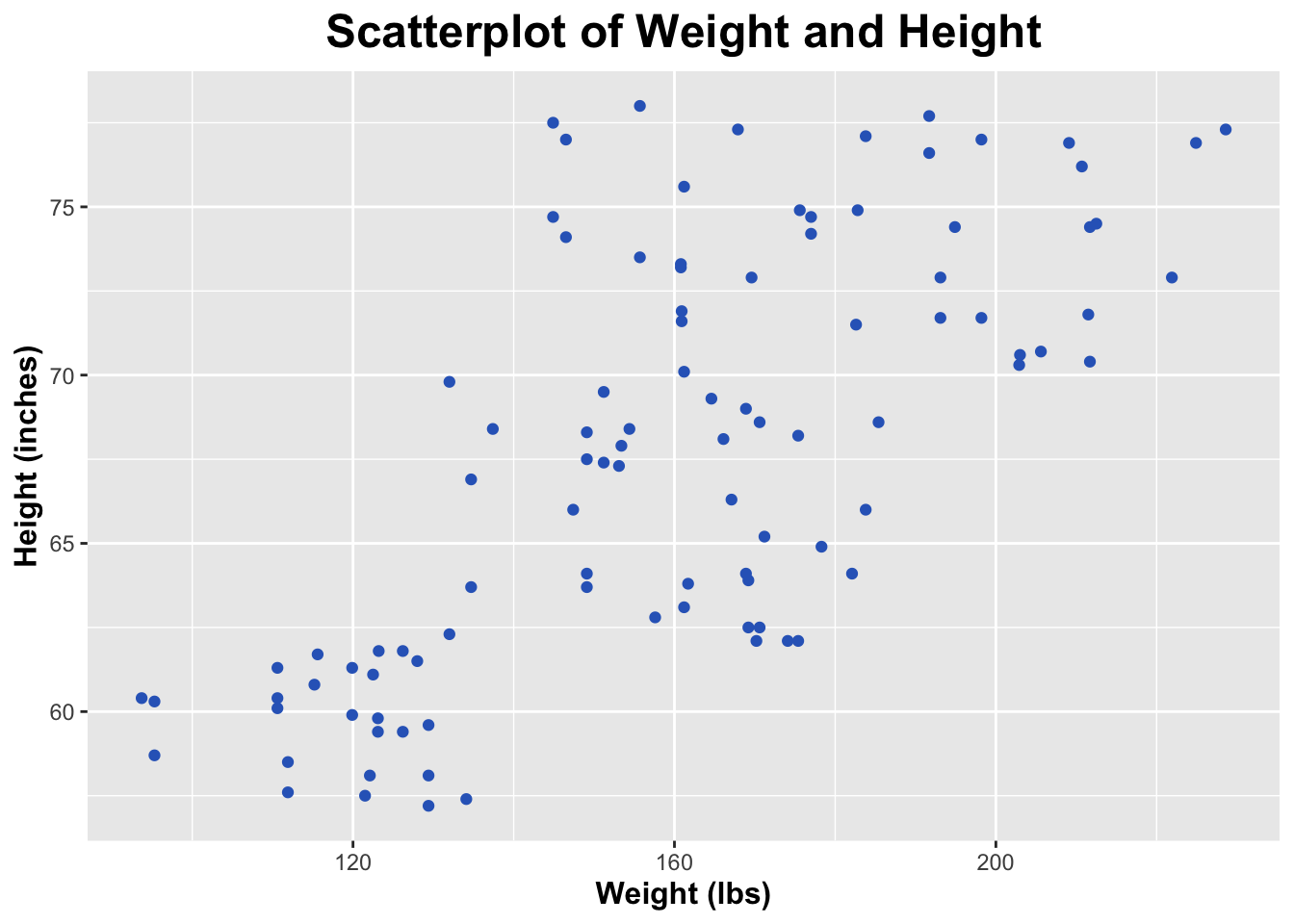
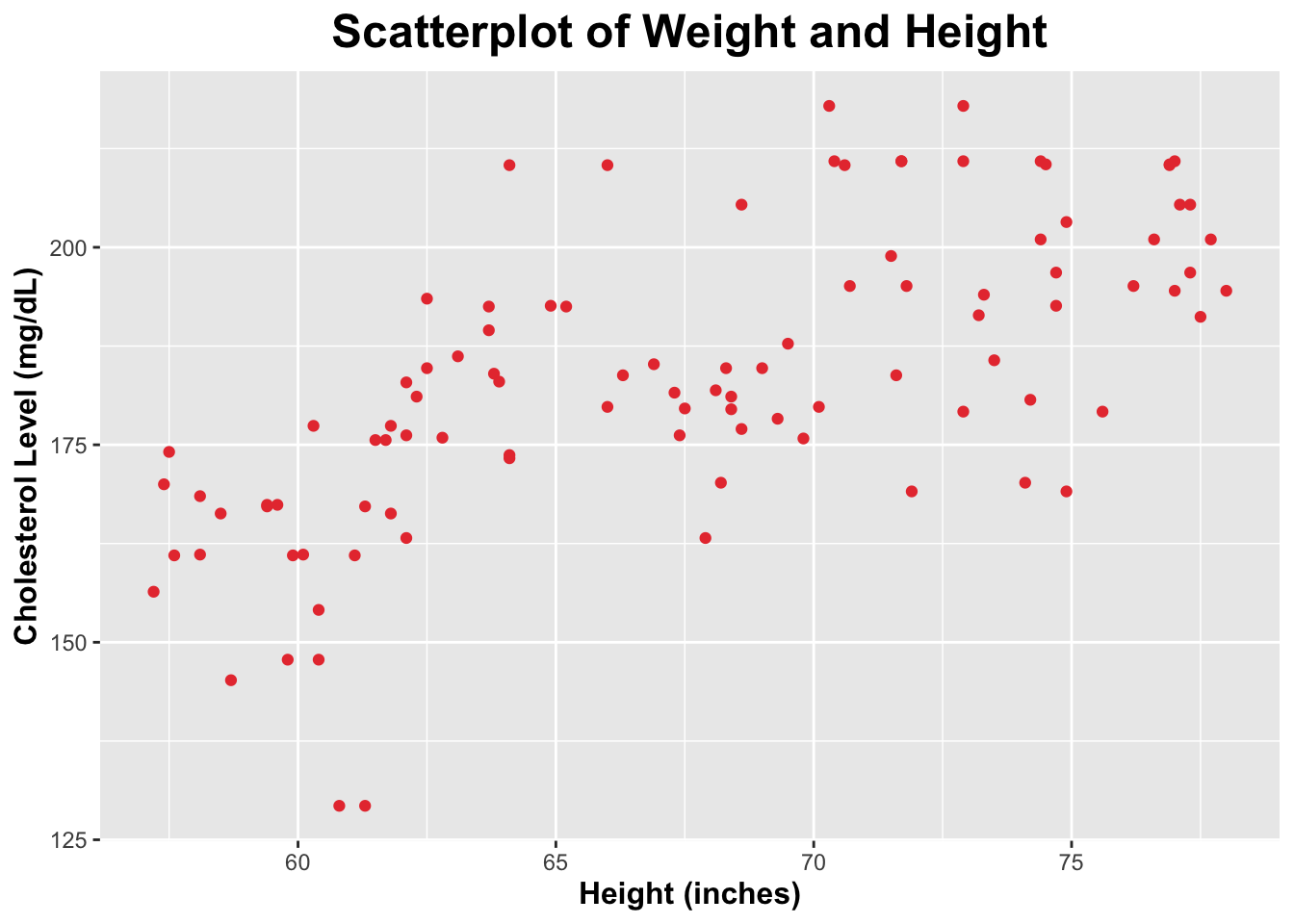
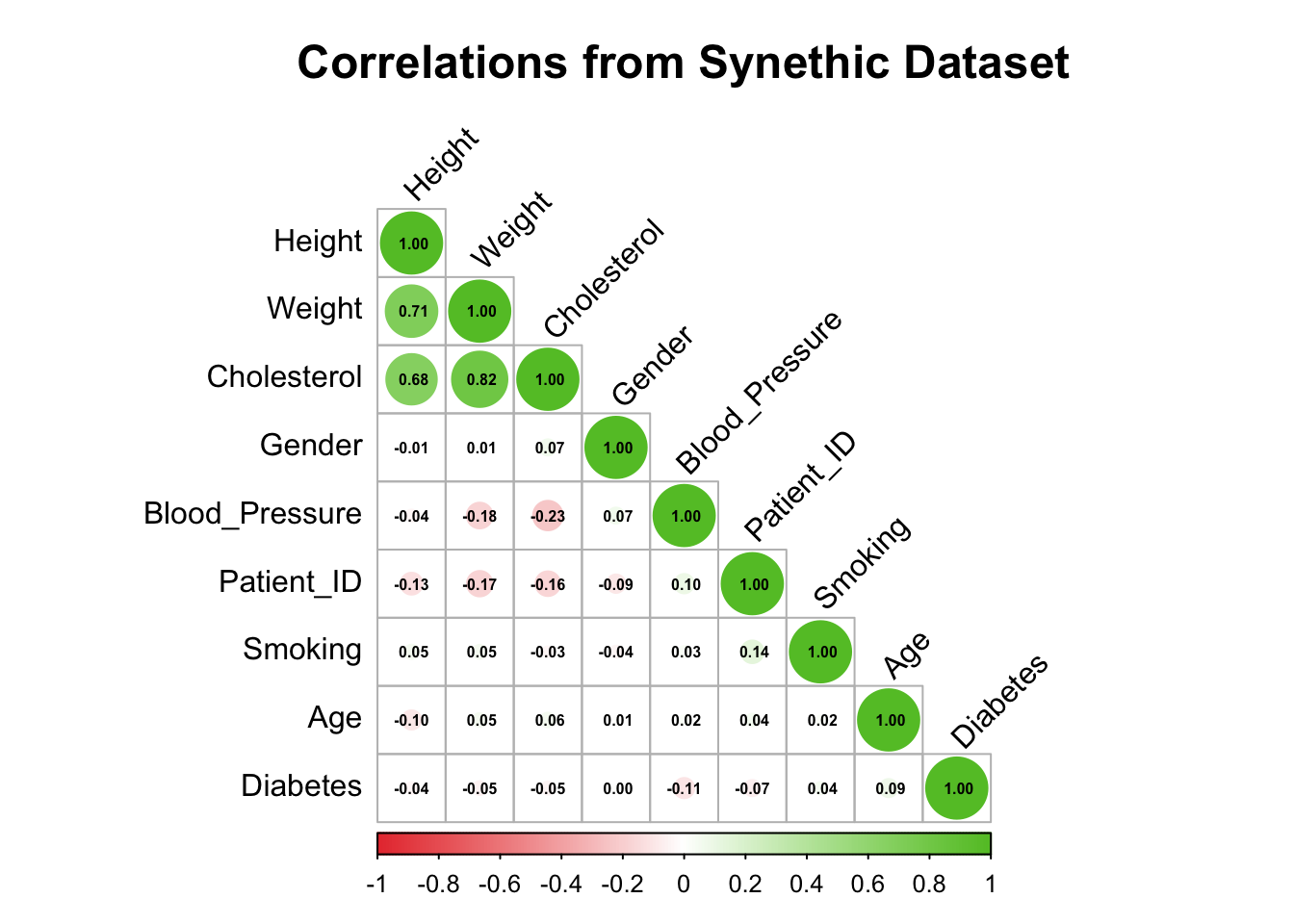
$ Height <dbl> 68.6, 69.0, 65.2, 78.0, 77.1, 62.1, 62.1, 71.5, 69.5, …
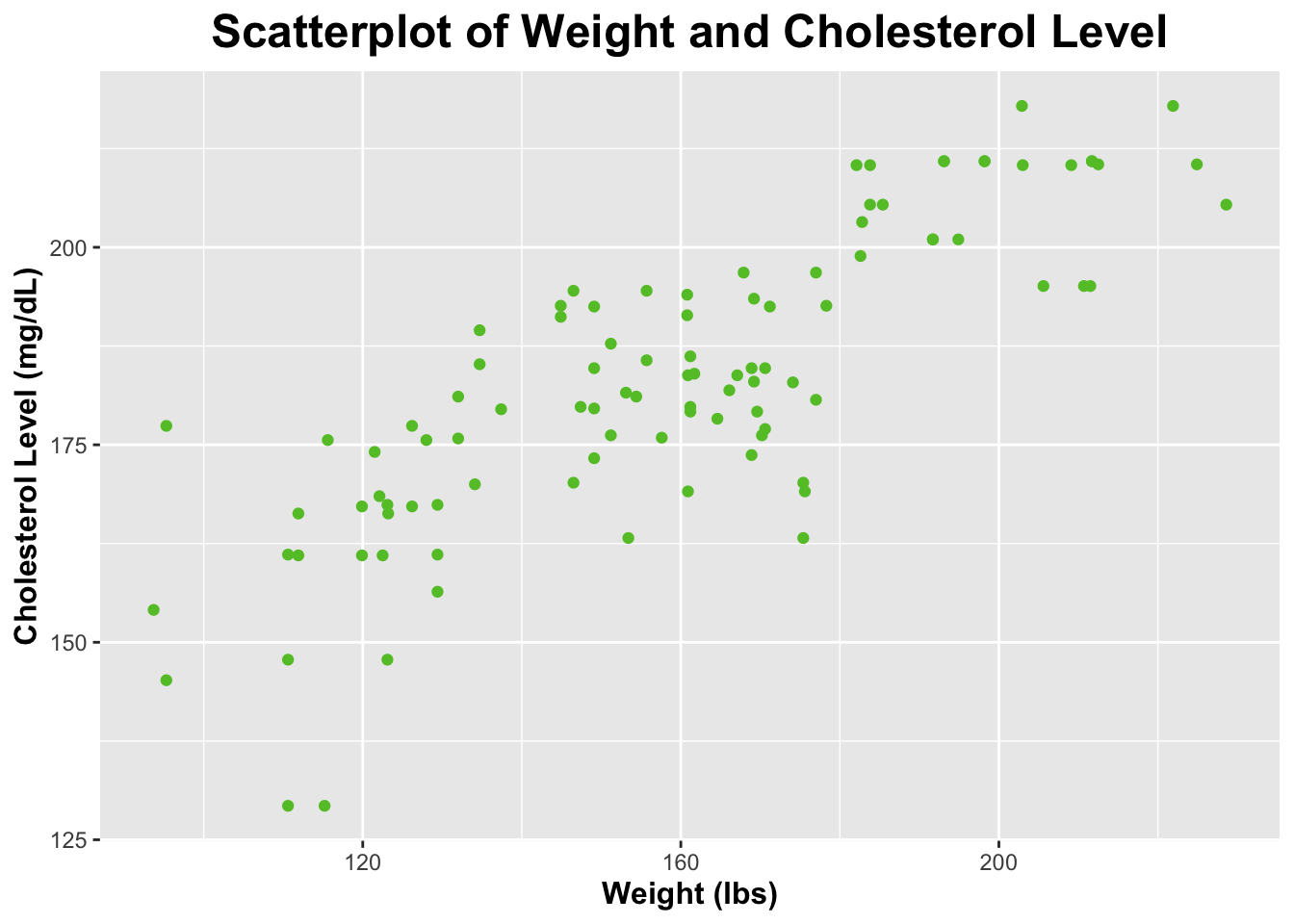
$ Weight <dbl> 170.6, 168.9, 171.2, 155.7, 183.8, 174.1, 175.4, 182.6…
$ Blood_Pressure <dbl> 96.0, 125.8, 156.6, 109.0, 100.8, 95.2, 155.1, 99.9, 1…
$ Cholesterol <dbl> 177.0, 184.7, 192.5, 194.5, 205.4, 182.9, 163.2, 198.9…
$ Diabetes <dbl> 1, 1, 1, 0, 0, 0, 0, 0, 1, 1, 1, 1, 0, 0, 1, 0, 0, 1, …
$ Smoking <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 1, 1, 1, 0, 1, 1, 1, 0, …